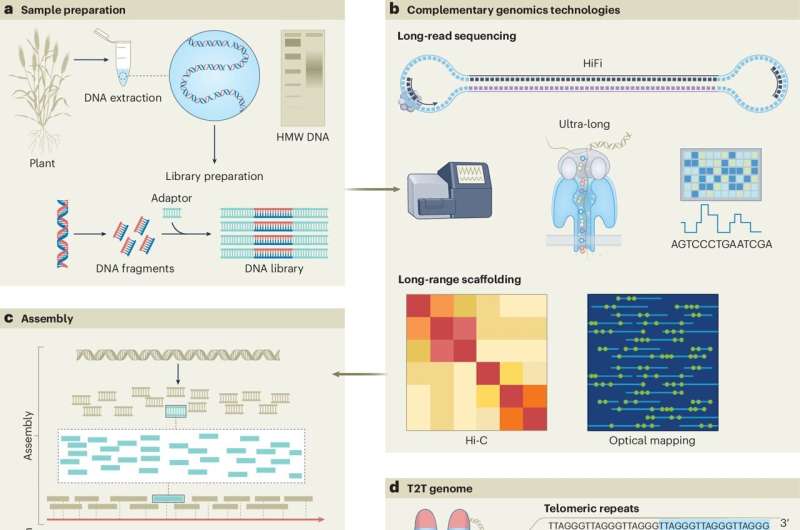
Summary of different strategies for developing T2T genome assemblies. Credit: Genetics of Nature (2024). DOI: 10.1038/s41588-024-01830-7
Entirely new crop varieties that can better withstand drought, salt and pests are within reach thanks to international genomic research published today in Genetics of Nature.
The study, “Unlocking plant genetics with telomere-to-telomere genome assemblies,” was led by researchers at Murdoch University’s Center for Crop and Food Innovation (CCFI).
Complete Telomere-to-Telomere (T2T) assemblies allow scientists to map an entire genome to support molecular breeding to improve plant performance under stress conditions.
Study leader and CCFI director Professor Rajeev Varshney said the genome assembly findings represented the future of genetics.
“Genome assembly is essentially like building a jigsaw, piece by piece. T2T genome assemblies provide a complete, end-to-end view of a genome in its entirety,” Professor Varshney said.
“Our job is to make sure that the thousands of genes within a genome are assembled in the right order, along the entire chromosome, and that each piece fits together perfectly.”
First author and CCFI researcher Dr. Vanika Garg, said the level of precision now possible in gene sequencing had led to the discovery.
“Until now, T2T genome assemblies have not been possible, and incomplete gene sequencing can lead to errors, such as mis-annotations,” said Dr. Garg.
“This means that the genes responsible for some desirable traits are lost. But thanks to advances in sequencing technologies, we can finally have T2T genome assemblies for every crop species.”
Professor Varshney and his research team are currently working in collaboration with several laboratories from Australia, UK, Germany, USA and China to develop T2T genome assemblies or near complete genome assemblies for several crops such as wheat, chickpea, beans , papaya, passion fruit. fruit, custard apple, banana and pineapple.
“Together with our national and international collaborators, we are applying T2T genome assembly methods to several crops and are extremely excited to see how this can advance agricultural research in Australia and overseas,” Professor Varshney said.
“T2T genome assemblies give access to difficult-to-sequence regions, which opens up a host of innovative research opportunities, such as creating entirely new crop varieties that meet our future needs.”
More information:
Vanika Garg et al, Unlocking plant genetics with telomere-telomere genome assemblies, Genetics of Nature (2024). DOI: 10.1038/s41588-024-01830-7
Provided by Murdoch University
citation: Telomere-to-telomere genome assembly research opens door to new crop varieties (2024, July 24) retrieved July 24, 2024 from https://phys.org/news/2024-07-telomere-genome-door -crop-varieties .html
This document is subject to copyright. Except for any fair agreement for study or private research purposes, no part may be reproduced without written permission. The content is provided for informational purposes only.
